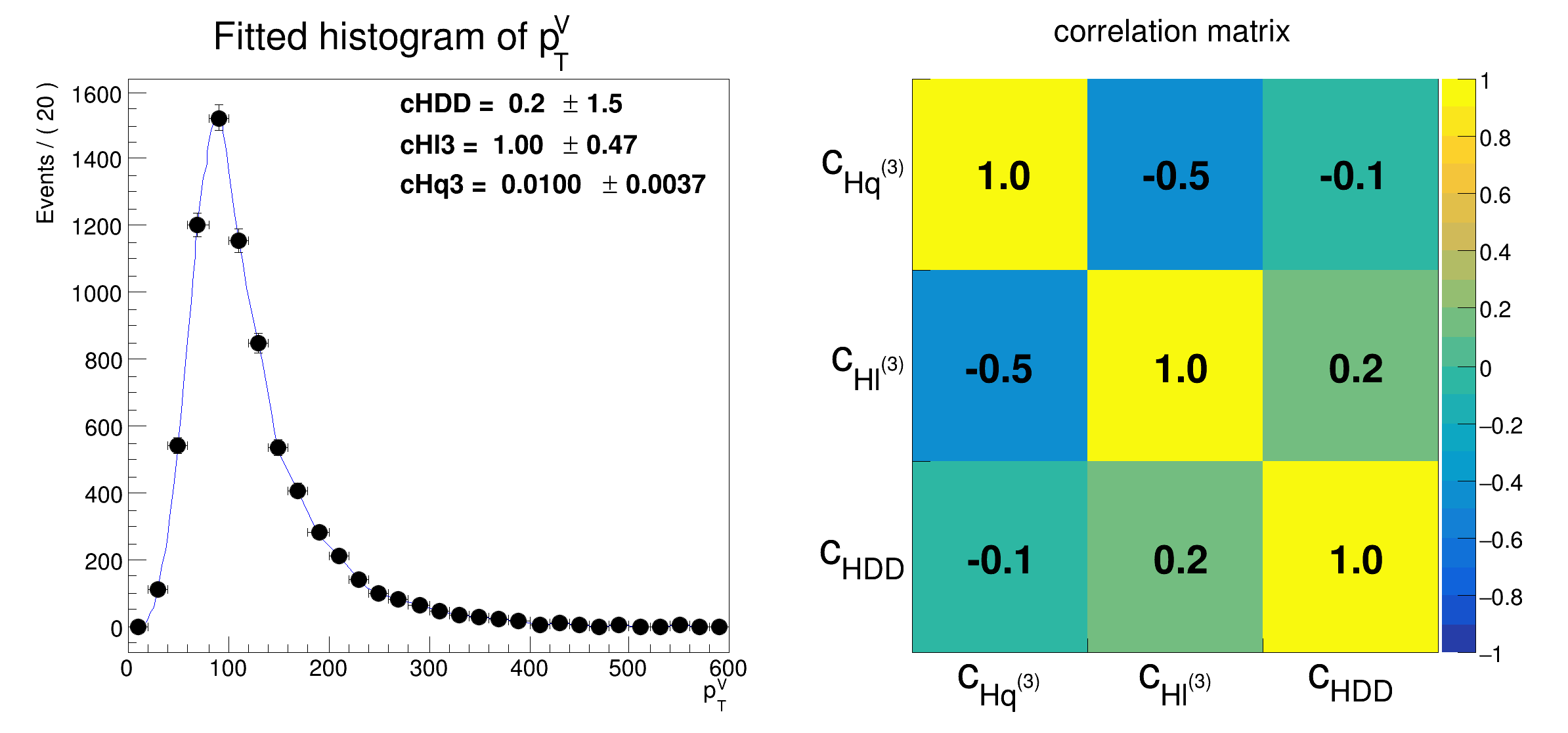
a morphing function is setup as a function of three variables and a fit is performed on a pseudo-dataset.
{
std::string observablename = "pTV";
RooRealVar obsvar(observablename.c_str(),
"observable of pTV", 10, 600);
RooRealVar cHq3(
"cHq3",
"EFT modifier", -10.0, 10.0);
cHq3.setAttribute("NewPhysics", true);
RooRealVar cHl3(
"cHl3",
"EFT modifier", -10.0, 10.0);
cHl3.setAttribute("NewPhysics", true);
RooRealVar cHDD(
"cHDD",
"EFT modifier", -10.0, 10.0);
cHDD.setAttribute("NewPhysics", true);
std::string infilename = std::string(
gROOT->GetTutorialDir()) +
"/roofit/input_histos_rf_lagrangianmorph.root";
std::vector<std::string> samplelist = {"SM_NPsq0", "cHq3_NPsq1", "cHq3_NPsq2", "cHl3_NPsq1",
"cHl3_NPsq2", "cHDD_NPsq1", "cHDD_NPsq2", "cHl3_cHDD_NPsq2",
"cHq3_cHDD_NPsq2", "cHl3_cHq3_NPsq2"};
morphfunc.setParameter("cHq3", 0.01);
morphfunc.setParameter("cHl3", 1.0);
morphfunc.setParameter("cHDD", 0.2);
auto pseudo_hist = morphfunc.createTH1("pseudo_hist");
morphfunc.setParameter("cHq3", 0.0);
morphfunc.setParameter("cHl3", 0.0);
morphfunc.setParameter("cHDD", 0.0);
cHq3.setError(0.1);
cHl3.setError(0.1);
cHDD.setError(0.1);
auto fitres = model.fitTo(*pseudo_dh, SumW2Error(
true), Optimize(
false), Save(),
PrintLevel(-1));
auto hcorr = fitres->correlationHist();
auto postfit_hist = morphfunc.createTH1("morphing_postfit_hist");
auto frame0 = obsvar.frame(Title("Fitted histogram of p_{T}^{V}"));
postfit_dh.plotOn(frame0, Name(
"postfit_dist"), DrawOption(
"C"), LineColor(
kBlue), DataError(
RooAbsData::None),
XErrorSize(0));
pseudo_dh->plotOn(frame0, Name("input"));
gPad->SetLeftMargin(0.15);
gPad->SetRightMargin(0.05);
model.paramOn(frame0, Layout(0.50, 0.75, 0.9), Parameters(config.
couplings));
frame0->GetXaxis()->SetTitle("p_{T}^{V}");
frame0->Draw();
gPad->SetLeftMargin(0.15);
gPad->SetRightMargin(0.15);
hcorr->SetMarkerSize(3.);
hcorr->SetTitle("correlation matrix");
hcorr->GetYaxis()->SetTitleOffset(1.4);
hcorr->GetYaxis()->SetLabelSize(0.1);
hcorr->GetXaxis()->SetLabelSize(0.1);
hcorr->GetYaxis()->SetBinLabel(1, "c_{HDD}");
hcorr->GetYaxis()->SetBinLabel(2, "c_{Hl^{(3)}}");
hcorr->GetYaxis()->SetBinLabel(3, "c_{Hq^{(3)}}");
hcorr->GetXaxis()->SetBinLabel(3, "c_{HDD}");
hcorr->GetXaxis()->SetBinLabel(2, "c_{Hl^{(3)}}");
hcorr->GetXaxis()->SetBinLabel(1, "c_{Hq^{(3)}}");
hcorr->Draw("colz text");
c1->SaveAs(
"rf712_lagrangianmorphfit.png");
}
R__EXTERN TStyle * gStyle
virtual bool add(const RooAbsArg &var, bool silent=false)
Add the specified argument to list.
RooArgList is a container object that can hold multiple RooAbsArg objects.
The RooDataHist is a container class to hold N-dimensional binned data.
Class RooLagrangianMorphing is a implementation of the method of Effective Lagrangian Morphing,...
RooRealVar represents a variable that can be changed from the outside.
The RooWrapperPdf is a class that can be used to convert a function into a PDF.
void SetOptStat(Int_t stat=1)
The type of information printed in the histogram statistics box can be selected via the parameter mod...
void SetPaintTextFormat(const char *format="g")
The namespace RooFit contains mostly switches that change the behaviour of functions of PDFs (or othe...
std::string observableName
std::vector< std::string > folderNames
[#0] PROGRESS:InputArguments -- initializing physics inputs from file /home/sftnight/build/workspace/root-makedoc-v628/rootspi/rdoc/src/v6-28-00-patches.build/tutorials/roofit/input_histos_rf_lagrangianmorph.root with object name(s) 'pTV'
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x7fff9de6d6b8
[#0] PROGRESS:Caching -- current storage has size 10
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(pseudo_dh): fit range of variable pTV expanded to nearest bin boundaries: [10,600] --> [0,600]
[#0] PROGRESS:Caching -- creating cache from getCache function for 0x5629bbbfc860
[#0] PROGRESS:Caching -- current storage has size 10
[#0] PROGRESS:ObjectHandling -- observable: pTV
[#0] PROGRESS:ObjectHandling -- binWidth: binWidth_pTV
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39615.1) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.223088 cHl3=2.02101 cHq3=0.00780325
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.223088 +/- 0.1,cHl3 = 2.02101 +/- 0.1,cHq3 = 0.00780325 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.041358), trying to recover @ inputFunction=morphfunc=-0.0413585
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0413585
p.d.f value is less than zero (-0.043016), trying to recover @ inputFunction=morphfunc=-0.043016
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.043016
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39615.1) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.56869 cHl3=4.96014 cHq3=0.0199009
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.56869 +/- 0.1,cHl3 = 4.96014 +/- 0.1,cHq3 = 0.0199009 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.074541), trying to recover @ inputFunction=morphfunc=-0.0745407
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0745407
p.d.f value is less than zero (-1.173323), trying to recover @ inputFunction=morphfunc=-1.17332
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.17332
p.d.f value is less than zero (-1.063175), trying to recover @ inputFunction=morphfunc=-1.06317
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.06317
p.d.f value is less than zero (-1.234351), trying to recover @ inputFunction=morphfunc=-1.23435
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.23435
p.d.f value is less than zero (-1.674381), trying to recover @ inputFunction=morphfunc=-1.67438
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-1.67438
p.d.f value is less than zero (-0.075079), trying to recover @ inputFunction=morphfunc=-0.0750793
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0750793
... (remaining 10 messages suppressed)
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39615.1) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.318765 cHl3=2.86725 cHq3=0.0111509
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.318765 +/- 0.1,cHl3 = 2.86725 +/- 0.1,cHq3 = 0.0111509 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.070498), trying to recover @ inputFunction=morphfunc=-0.0704979
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0704979
p.d.f value is less than zero (-0.063999), trying to recover @ inputFunction=morphfunc=-0.0639991
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0639991
p.d.f value is less than zero (-0.140607), trying to recover @ inputFunction=morphfunc=-0.140607
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.140607
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39615.1) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.193718 cHl3=1.7579 cHq3=0.00677582
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.193718 +/- 0.1,cHl3 = 1.7579 +/- 0.1,cHq3 = 0.00677582 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.029110), trying to recover @ inputFunction=morphfunc=-0.0291104
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0291104
p.d.f value is less than zero (-0.011532), trying to recover @ inputFunction=morphfunc=-0.0115319
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0115319
RooAbsMinimizerFcn: Minimized function has error status.
Returning maximum FCN so far (39615.1) to force MIGRAD to back out of this region. Error log follows.
Parameter values: cHDD=0.820784 cHl3=1.67112 cHq3=0.0140305
RooNLLVar::nll_wrap_pdf_pseudo_dh[ parameters=(binWidth_pTV,cHDD,cHl3,cHq3,kSM,nNP0,nNP1,nNP2,nNP3,nNP4) ]
function value is NAN @ parameters=(binWidth_pTV = 0.05,cHDD = 0.820784 +/- 0.1,cHl3 = 1.67112 +/- 0.1,cHq3 = 0.0140305 +/- 0.1,kSM = 1,nNP0 = 1,nNP1 = 1,nNP2 = 1,nNP3 = 1,nNP4 = 1)
RooWrapperPdf::wrap_pdf[ inputFunction=morphfunc ]
p.d.f value is less than zero (-0.010720), trying to recover @ inputFunction=morphfunc=-0.0107196
getLogVal() top-level p.d.f evaluates to NaN @ inputFunction=morphfunc=-0.0107196
[#1] INFO:Fitting -- RooAbsPdf::fitTo(wrap_pdf) Calculating sum-of-weights-squared correction matrix for covariance matrix
[#1] INFO:DataHandling -- RooDataHist::adjustBinning(morphing_postfit_dh): fit range of variable pTV expanded to nearest bin boundaries: [0,600] --> [0,600]
[#1] INFO:InputArguments -- RooAbsData::plotOn(pseudo_dh) INFO: dataset has non-integer weights, auto-selecting SumW2 errors instead of Poisson errors
[#1] INFO:Plotting -- RooPlot::updateFitRangeNorm: New event count of 7389.24 will supercede previous event count of 7396.07 for normalization of PDF projections